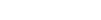
Researchers have used data from experiments conducted in human and worm models to explore the mutational causes of cancer and identify possible routes for future treatments.
The team consists of scientists from the European Bioinformatic Institute (EMBL-EBI), the University of Dundee and the Wellcome Sanger Institute. Results from the study were published in Genome Research, demonstrating the controlled experiments conducted on the nematode worm C. elegans to be applicable for humans.

Discover B2B Marketing That Performs
Combine business intelligence and editorial excellence to reach engaged professionals across 36 leading media platforms.
Understanding the DNA mutational processes that lead to cancer could shed new light on cancer causes, and thus possible new treatment routes.
Such DNA mutations are catalysed by factors including UV radiation and smoking, as well as natural errors during cell division. Though the body identifies and repairs these mutations, it is an imperfect correction which often leaves some untreated in the body.
“We know that cancer is caused by alterations in the DNA, and that tumour cells are genetically different from the normal ones,” EMBL-EBI PhD student Nadia Volkova told Drug Development Technology.
“However, when we look at the somatic mutations in a cancer, we only see the result of a complex combination of mutational processes that have shaped this particular tumour. Linking an observed pattern to an underlying biological deficiency is very hard, since the same process can produce different damage depending on the genetic environment.
US Tariffs are shifting - will you react or anticipate?
Don’t let policy changes catch you off guard. Stay proactive with real-time data and expert analysis.
By GlobalData“By disentangling which mutational processes were active in a tumour, and especially which of them are active now, it is possible to enhance the knowledge of the aetiology and biochemistry of a tumour.”
Previous studies have demonstrated DNA repair pathway mismatch repair (MMR) to be particularly associated with an increased risk of cancer. The recent research examined the MMR pathway using C. elegans models.
“C. elegans has short generation time, it’s self-fertilizing, it has a shorter and simpler genome, and most of its DNA repair mechanisms are conserved between C. elegans and humans,” said Volkova.
“As the goal of the main project was to generate a panel of samples with various DNA repair deficiencies to study the mutagenesis under different conditions, C. elegans seemed to be a good candidate for it.”
Dr Bettina Meier from the Dundee Centre for Gene Regulation and Expression initiated the study through examining mutations that result from a C. elegans defect in a single DNA repair pathway.
“As it only takes three days to propagate these worms from one generation to the next, the process of studying how DNA is passed on is greatly expedited,” Dundee Centre for Gene Regulation and Expression principal investigator Professor Anton Gartner said.
“DNA mismatch repair is propagated for many generations and this allowed us to deduce a distinct mutational pattern. The big question was if the same type of mutagenesis also occurred in human cancer cells.”
In response to this question, Volkova compared data from the C. elegans models with that from 500 human cancer genomes.
“We found a resemblance between the most common signature associated with mutations in MMR genes in humans and the patterns found in nematode worms,” Volkova said.
“This suggests that the same mutational process operates in nematodes and humans. Our approach allows us to find the exact profile of MMR deficiency and to understand more about what happens when DNA repair goes wrong.”
The project is part of a larger consortium studying mutational patterns of DNA repair deficiencies and carcinogens through model organisms. Volkova told Drug Development
“The ultimate goal is to generate a large catalogue of mutational signatures which will help to refine and explain the mutational signatures in human cancers.”